| 33 | ||
| 32 | ||
| 31 | ||
| 30 | 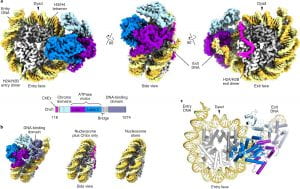 | Nodelman, I.M, Das, S., Faustino, A.M., Fried, S.D., Bowman, G.D., Armache, J.-P. (2022). Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state NAT. STRUCT. MOL. BIOL. FEB 16;29(2):121-129. PMID: 35173352 |
| 29 |  | Patterson, D.C., Liu, Y., Das, S. , Yennawar, N.H., Armache, J.-P., Kincaid J.R., and Weinert, E.E. (2021). Heme-Edge Residues Modulate Signal Transduction within a Bifunctional Homo-Dimeric Sensor Protein Biochemistry, DEC 2021, 60(49):3801-3812. PMID: 34843212 |
| 28 | 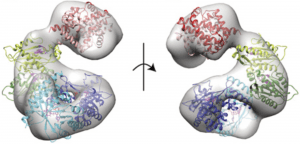 | Patterson, D.C., Perez Ruiz, M. , Yoon, H., Walker, J.A., Armache, J.-P., Yennawar, N.H., and Weinert, E.E. (2021). Differential ligand-selective control of opposing enzymatic activities within a bifunctional c-di-GMP enzyme PNAS, SEP 2021, 118 (36). PMID: 34475207 |
| 27 |  | Review Wilson, S.L., Way, G.P., Bittremieux, W., Armache, J.-P., Haendel, M.A., Hoffman, M.M. (2021). Sharing biological data: why, when, and how FEBS Letters, APR 11 2021, PMID: 33843054 |
| 26 | 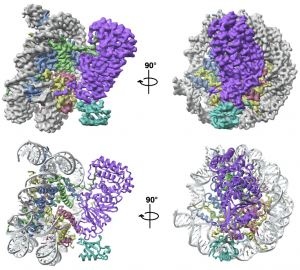 | Valencia-Sanchez, M.I., De Ioannes, P., Wang, M., Truong, D.M., Lee, R., Armache, J.-P., Boeke, J.D. and Armache, K.-J. (2021). Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. SCIENCE, JAN 22; 371 (6527). PMID: 33479126 |
| 25 | 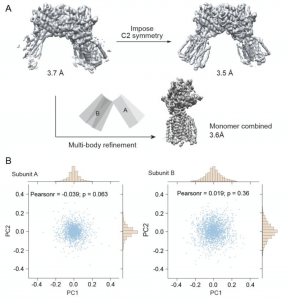 | Review Armache, J.-P., Cheng, Y (2019). Single particle cryo-EM-beyond the resolution. National Science Review |
| 24 | 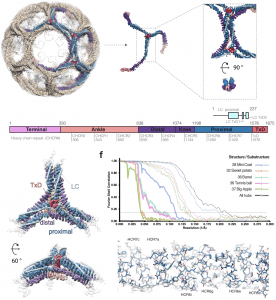 | Morris KL, Jones JR, Halebian M, Wu S, Baker M, Armache J.-P.., Avila Ibarra A, Sessions RB, Cameron AD, Cheng Y, Smith CJ. (2019). Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly. NAT. STRUCT. MOL. BIOL., Oct;26(10):890-898. PMID: 31582853 |
| 23 | 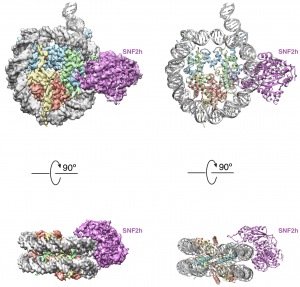 | Armache, J.-P.*, Gamarra, N.*, Johnson, S.L., Wu, S., Leonard, J., Narlikar, G. and Cheng, Y. (2019). Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. ELIFE. JUN 18; 8. (* equal contribution). PMID: 31210637 |
| 22 | 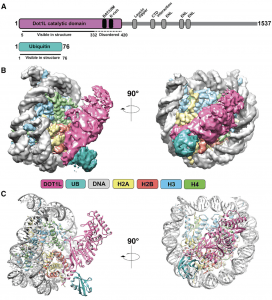 | Valencia-Sanchez, M.I., De Ioannes, P., Wang,M., Vasilyev,N. Chen, R. Nudler, E., Armache, J.-P. and Armache, K.-J. (2019). Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. MOLECULAR CELL BIOLOGY, JUN 6; 74(5):1010-1019. PMID: 30981630 |
| 21 | 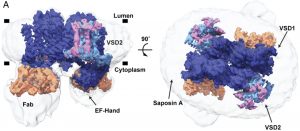 | Kintzer, A.F., Green, E., Dominik, P., Bridges, M., Armache, J.-P., Deneka, D., Kim, S., Hubbell, W., Kossiakoff, A., Cheng, Y., Stroud, R. (2018). The structural basis for activation of voltage sensor domains in an ion channel TPC1. PROC. NATL. ACAD. SCI. U.S.A., SEP 25;115. PMID: 30190435 |
| 20 |  | REVIEW Heymann, J.B., Marabini, R., Kazemi, M., Sorzano, C.O.S., Holmdahl, M., Mendez, J.H., Stagg, S.M., Jonic, S., Palovcak, E., Armache, J.-P., Zhao, J., Cheng, Y., Pintilie, G., Chiu, W., Patwardhan, A., Carazo, J.M. (2018). The first single particle analysis Map Challenge: A summary of the assessments. J. STRUCT. BIOL., NOV; 204(2):291-300. PMID: 30114512 |
| 19 | 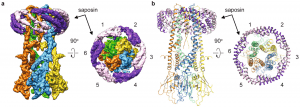 | Nguyen, N.X., Armache, J.-P., Lee, C., Yang, Y., Zeng, W., Mootha, V.K., Cheng, Y., Bai, X.-C., Jiang, Y. (2018). Cryo-EM structure of the mitochondrial calcium uniporter. NATURE, JULY:1-19. PMID: 29995855 |
| 18 | 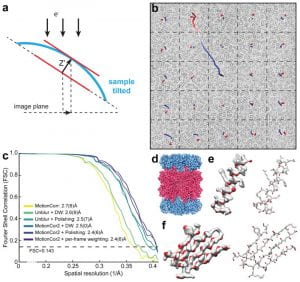 | Zheng, S.Q., Palovcak, E., Armache, J.-P., Verba, K.A., Cheng, Y. and Agard, D.A. (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. NATURE METHODS, APR;14(4):331-332. PMID: 28250466 |
| 17 | 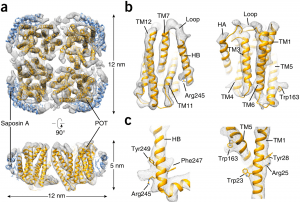 | Frauenfeld J., Löving R., Armache J.-P., Sonnen, A.F.-P., Guettou, F., Moberg, P., Zhu, L., Jegerschöld, C., Flayhan, A., Briggs, J.A.G., Garoff, H., Löw, C., Cheng, Y. & Nordlund, P. A (2016). A saposin-lipoprotein nanoparticle system for membrane proteins. NATURE METHODS, MARCH 1-11. PMID: 26950744 |
| 16 | 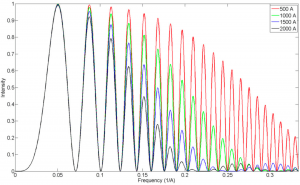 | Review Wu S., Armache J.-P., Cheng Y. (2016). Single-particle cryo-EM data acquisition by using direct electron detection camera. MICROSCOPY, 65(1), 35-41. PMID: 26546989 |
| 15 | 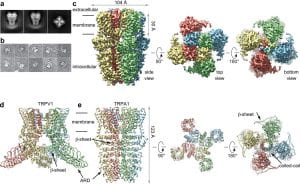 | Paulsen, C. E.*, Armache J.-P.*, Gao Y., Cheng Y., Julius D. (2015). Structure of the TRPA1 ion channel suggests regulatory mechanisms. NATURE, 520 (7548), 511-517. (* equal contribution) PMID: 25855297 |
| 14 | 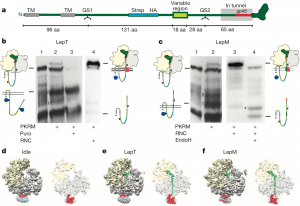 | Gogala, M., Becker, T., Beatrix, B., Armache, J.-P., Barrio-Garcia, C., Berninghausen, O., Beckmann, R. (2014). Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. NATURE, 506, 107-110. PMID: 24499919 |
| 13 |  | Anger, A.M.*, Armache, J.-P.*, Berninghausen, O., Habeck, M., Subklewe, M., Wilson, D.N., and Beckmann, R. (2013). Structures of the human and Drosophila 80S ribosome. NATURE, 497, 80–85. (* equal contribution) PMID: 23636399 |
| 12 |  | Armache, J.-P.*, Anger, A.M.*, Márquez, V., Franckenberg, S., Fröhlich, T., Villa, E., Berninghausen, O., Thomm, M., Arnold, G.J., Beckmann, R., and Wilson, D.N. (2013). Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. NUCLEIC ACIDS RES., 41, 1284–1293. (* equal contribution) PMID: 23222135 |
| 11 |  | Grela, P., Gajda, M.J., Armache, J.-P., Beckmann, R., Krokowski, D., Svergun, D.I., Grankowski, N., and Tchórzewski, M. (2012). Solution structure of the natively assembled yeast ribosomal stalk determined by small-angle X-ray scattering. BIOCHEM. J., 444, 205–209. PMID: 22458705 |
| 10 |  | Becker, T., Franckenberg, S., Wickles, S., Shoemaker, C.J., Anger, A.M., Armache, J.-P., Sieber, H., Ungewickell, C., Berninghausen, O., Daberkow, I., Karcher, A., Thomm, M. Hopfner, K.-P., Green, R., and Beckmann R. (2012). Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. NATURE, 482, 501–506. PMID: 22358840 |
| 9 | 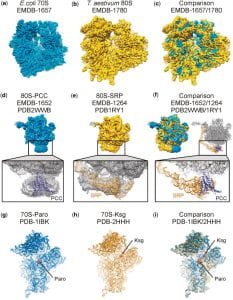 | Jarasch, A., Dziuk, P., Becker, T., Armache, J.-P., Hauser, A., Wilson, D.N., and Beckmann, R. (2012). The DARC site: a database of aligned ribosomal complexes. NUCLEIC ACIDS RES., 40, D495–D500. PMID: 22009674 |
| 8 | 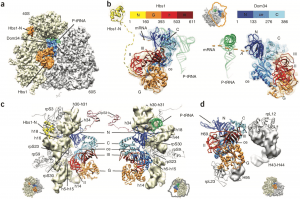 | Becker, T., Armache, J.-P., Jarasch, A., Anger, A.M., Villa, E., Sieber, H., Motaal, B.A., Mielke, T., Berninghausen, O., and Beckmann, R. (2011). Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. NAT. STRUCT. MOL. BIOL., 18, 715–720. PMID: 21623367 |
| 7 | 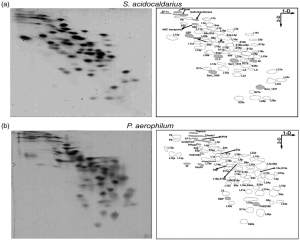 | Márquez, V., Fröhlich, T., Armache, J.-P., Sohmen, D., Dönhöfer, A., Mikolajka, A., Berninghausen, O., Thomm, M., Beckmann, R., Arnold, G.J., and Wilson, D.N. (2011). Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. J. MOL. BIOL., 405, 1215–1232. PMID: 21134383 |
| 6 | 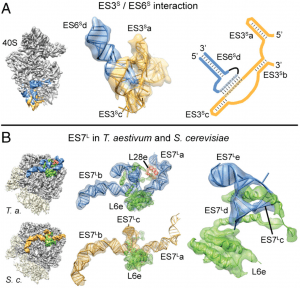 | Armache, J.-P.*, Jarasch, A.*, Anger, A.M.*, Villa, E., Becker, T., Bhushan, S., Jossinet, F., Habeck, M., Dindar, G., Franckenberg, S., Márquez, V., Mielke, T., Thomm, M., Berninghausen, O., Beatrix, B., Söding, J., Westhof, E., Wilson, D.N., and Beckmann, R. (2010a). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. PROC. NATL. ACAD. SCI. U.S.A., 107, 19748–19753. (* equal contribution) PMID: 20980660 |
| 5 | 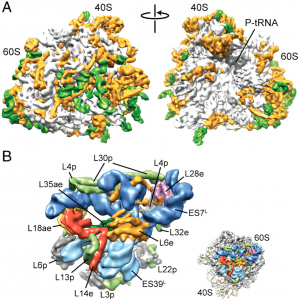 | Armache, J.-P.*, Jarasch, A.*, Anger, A.M.*, Villa, E., Becker, T., Bhushan, S., Jossinet, F., Habeck, M., Dindar, G., Franckenberg, S., Márquez, V., Mielke, T., Thomm, M., Berninghausen, O., Beatrix, B., Söding, J., Westhof, E., Wilson, D.N., and Beckmann, R. (2010b). Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. PROC. NATL. ACAD. SCI. U.S.A., 107, 19754–19759. (* equal contribution) PMID: 20974910 |
| 4 | 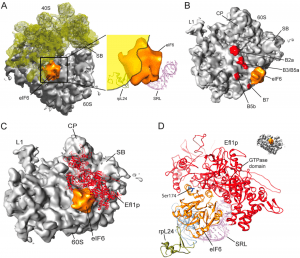 | Gartmann, M., Blau, M., Armache, J.-P., Mielke, T., Topf, M., and Beckmann, R. (2010). Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J. BIOL. CHEM., 285, 14848–14851. PMID: 20356839 |
| 3 | 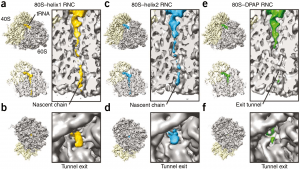 | Bhushan, S., Gartmann, M., Halic, M., Armache, J.-P., Jarasch, A., Mielke, T., Berninghausen, O., Wilson, D.N., and Beckmann, R. (2010). alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. NAT. STRUCT. MOL. BIOL., 17, 313–317. PMID: 20139981 |
| 2 | 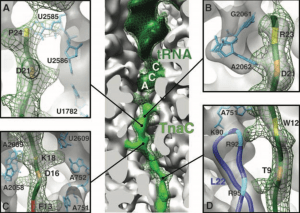 | Becker, T., Bhushan, S., Jarasch, A., Armache, J.-P., Funes, S., Jossinet, F., Gumbart, J., Mielke, T., Berninghausen, O., Schulten, K., Westhof, E., Gilmore, R., Mandon, E.C, and Beckmann R. (2009). Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. SCIENCE, 326, 1369–1373. PMID: 19933108 |
| 1 |  | Seidelt, B., Innis, C.A., Wilson, D.N., Gartmann, M., Armache, J.-P., Villa, E., Trabuco, L.G., Becker, T., Mielke, T., Schulten, K., Steitz, T.A., and Beckmann R. (2009). Structural insight into nascent polypeptide chain-mediated translational stalling. SCIENCE, 326, 1412–1415. PMID: 19933110 |